- History & Evolution
- Biosynthesis and dietary uptake
- 3-IAA and the gut microbiome
- 3-IAA and cancer
- 3-IAA as a biomarker
- Potential dietary and microbiome-based interventions with 3-IAA
- References
History & Evolution
1925: first synthesized (Majima et al., 1925) | 1933: identified as plant hormone (Weissbach et al., 1959) | 2020: identified as potential mediator of AhR activation (Sadik et al., 2020)
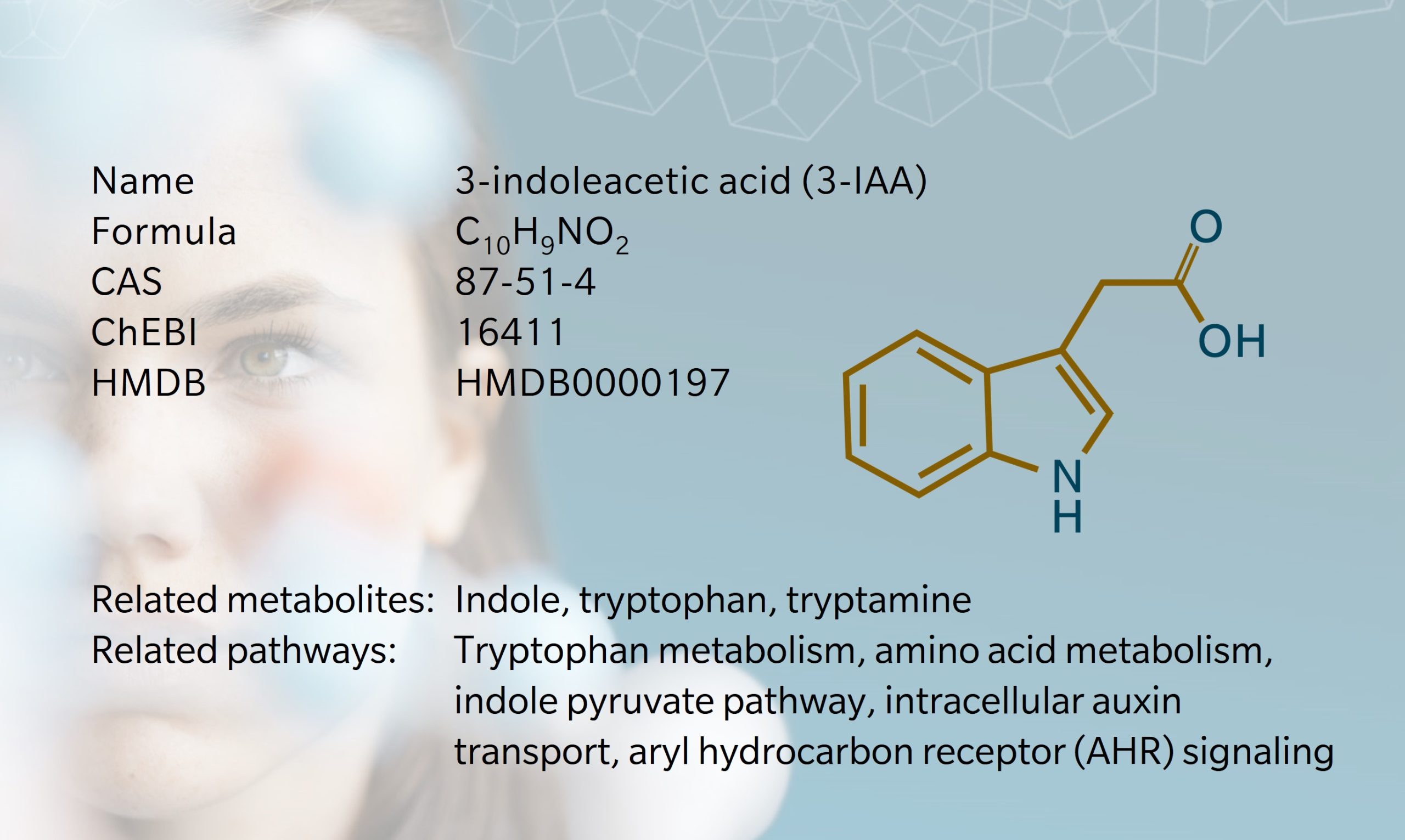
Indole-3-acetic acid, also referred to as 3-indoleacetic acid (3-IAA), is known as the most common plant growth hormone, though it is also found in mammals (Jones et al., 1995). Our understanding of it comes primarily from its role in plant biology, but interestingly, its function as a phytohormone was actually determined using human urine samples, in 1933 (Weissbach et al., 1959).
As a microbial metabolite of tryptophan, 3-IAA is associated with several important biological processes including immune function, inflammation and metabolic homeostasis. There’s a growing body of evidence linking indole derivatives with digestive disorders and toxicity (Zhang et al., 2020). Much current research focuses on 3-IAA’s role in activating the aryl hydrocarbon receptor (AhR) pathway, which presents potential therapeutic options for cancer and autoimmune disease (Shen et al., 2022).
Advances in omics techniques are allowing us to learn more about this metabolite. Researchers can investigate 3-IAA and other indole derivatives as potential biomarkers and mediators of disease, in a way they couldn’t before. This is hugely valuable given the prevalence of diet- and lifestyle-related disease and the role of the microbiome in those diseases.
An interesting recent discovery using omics techniques highlights the role of tryptophan-derived indole alkaloids in cellular responses to environmental change (Chen et al., 2023) . Prenyl indole alkaloids biosynthesized from tryptophan have been found to dramatically accumulate in the fungus Thermomyces dupontii under cold stress. Metabolomics revealed 3-IAA to be among the enriched metabolites, suggesting its involvement in a lipid-mediated fungal response to cold.
Biosynthesis vs. dietary uptake

In mammals, 3-IAA’s precursor indole is metabolized from tryptophan by gut microbiota. Tryptophan is an essential amino acid, which means 3-IAA production largely depends on dietary sources of tryptophan – typically dairy, eggs and meat. As these dietary proteins break down, tryptophan is released and converted into catabolites including indole, tryptamine and kynurenine.
Read our metabolite of the month article on tryptophan.
The vast majority of ingested tryptophan is metabolized to kynurenine in the gut by local immune cells and intestinal epithelial cells expressing the enzyme indoleamine 2,3-dioxygenase 1 (IDO1). An estimated 5% of the tryptophan pool is metabolized by microbial tryptophanase and decarboxylase in the indole pathway, and an even smaller fraction is converted to serotonin and melatonin (Taleb, 2019; Wyatt et al., 2021).
3-IAA can be synthesized from several precusors depending on the enzymatic capacities of the intestinal microbiome. Indole pyruvic acid, indole acetamide and indole acetaldehyde are commonly reported as precursors of 3-IAA (Hendrikx et al., 2019; Taleb, 2019; Wyatt et al., 2021). Small amounts of indole are also metabolized into the uremic toxin indoxyl sulfate.
In addition to exogenous biosynthesis, 3-IAA may also be produced endogenously from tryptophan. For instance, cells transfected with the gene for interleukin-4-induced 1 (IL4I1) metabolized tryptophan to indole-3-pyruvic acid and then to 3-IAA (Zhang et al., 2020).
As a side note, this pathway also involves the decarboxylation of indole acetic acid into skatole, the intestinal metabolite 3-methylindole that sometimes causes an off-flavor in pork from male pigs (Roager et al., 2018).
3-IAA and the gut microbiome
Most of the reactions involved in 3-IAA synthesis occur in the small intestine and colon, with the resulting metabolites entering the blood circulation through the intestinal epithelium (Riazati, N. et al., 2022). In the liver, 3-IAA can combine with glutamine to produce indole acetyl glutamine, or be metabolized by peroxidases to indole-3-aldehyde (Gao, 2018; De Mello et al., 1980).
The synthesis of 3-IAA from dietary tryptophan is particularly associated with enzymatic reactions involving the Clostridium and Bacteriodes species of intestinal bacteria (Russell et al., 2013; Gao, 2018).
In humans, 3-IAA has been detected in blood, feces, urine, saliva and cerebrospinal fluid (Shen et al., 2022) (HMDB). Fecal samples of healthy adult humans contain around 5 µM of 3-IAA (Lamas et al., 2016). Serum concentrations are closer to 1 µM (Rosas, H. et al., 2015). Both blood and urinary levels vary greatly from individual to individual, possibly due to microbiome differences (Rosas et al., 2015; Pavlova et al., 2017).
There is a strong correlation between the gut microbiota and tryptophan metabolism, suggesting that changes in the composition of the gut microbiome could influence tryptophan levels and consequently 3-IAA production (Roager et al., 2018). These findings suggest that manipulating gut microbiota may be a therapeutic possibility for the treatment of many diseases associated with tryptophan metabolism and more specifically, with 3-IAA production.
3-IAA and cancer
Several studies show links between 3-IAA and cancer. Metabolomic analysis has been an invaluable tool in understanding these links and exploring potential treatment options.
For example, a 2023 study by Tintelnot et al. used metagenomic sequencing and targeted metabolomics to investigate factors that could shed light on the poor response to chemotherapy in patients with pancreatic ductal adenocarcinoma (PDAC). Patients who responded were found to have higher levels of 3-IAA. Mouse models showed that treatment with fecal microbiota transplantation, short-term dietary manipulation of tryptophan and oral 3-IAA administration increased the efficacy of chemotherapy (Tintelnot et al., 2023).
Watch the webinar: Joseph Tintelnot, PhD, joins biocrates to present the team’s research and discuss the potential for nutritional interventions in PDAC treatment.
Targeted metabolomic profiling was also used in a 2021 study of prognostic biomarkers in epithelial ovarian cancer (Hishinuma et al., 2021). This showed a correlation between IDO activity and disease risk, and found 3-IAA synthesis to be decreased in tumor-bearing patients. This suggests that IDO inhibitors may be a therapeutic option for patients with increased IDO activity.
In a 2017 study, the biocrates AbsoluteIDQ®p180 kit was used for the quantitative analysis of tryptophan and phenylalanine metabolites in patients with gastric carcinogenesis (Lario et al., 2017). The results showed significant alterations in tryptophan metabolism in patients with the disease, raising the possibility of biomarker candidates from metabolites in this pathway.
3-IAA could also be relevant for cancer therapy due to its cytotoxic effects when oxidized by horseradish peroxidase (HRP) (Folkes et al., 2001). Because 3-IAA is only toxic after oxidized decarboxylation, 3-IAA and HRP could be used as the basis for targeted cancer therapy. By targeting HRP to a tumor, 3-IAA could be activated in the tumor while minimizing systemic toxicity.
3-IAA as a biomarker
As well as being a promising biomarker for cancers, 3-IAA may be a useful diagnostic tool in other diseases, evidenced using metabolomic analysis. For example, males with long-term overweight and individuals with overweight from childhood to adolescence have been found to have lower levels of urinary 3-IAA (Oluwagbemigun et al., 2020). Indole metabolites may therefore act as indicators of cardiometabolic disease risk.
Indoleacetic acid has also been discussed as a marker of renal transporter-mediated drug-drug interactions (Gessner et al., 2023). A cross-sectional study found that higher levels of serum 3-IAA were significantly associated with knee pain scores in patients with osteoarthritis, suggesting its use as a pain marker (Mehta et al., 2023).
Plasma concentrations of 3-IAA have been explored as markers of systemic inflammation and immune activation, given the metabolite’s role in regulating immune activity via the AhR pathway (Riazati et al., 2022). As a ligand of AhR and free radical scavenger, 3-IAA mediates toxicity and inflammation, both limiting production of pro-inflammatory cytokines and modulating the immune cells that trigger anti-inflammatory cytokine production (Krishnan et al., 2018).
Potential dietary and microbiome-based interventions with 3-IAA
The links between diet, the metabolome and disease risk are well-established. Could dietary or microbiome-focused interventions be used to modify 3-IAA concentrations? Could doing so help mitigate disease risk or influence treatment efficacy? Research suggests this may be the case.
A high-fiber-low-protein diet has been found to favor microbial production of 3-IAA (Huang et al., 2023). High-fat diets are associated with lower indole production (Hou et al., 2023). Total protein intake is positively associated with plasma 3-IAA, suggesting that dietary tryptophan may influence health via the microbiome (Riazati et al., 2022).
Patients with inflammatory bowel disease (IBD) have been found to have reduced fecal concentrations of 3-IAA, and in mice, oral administration of indole and indole derivatives appeared to mediate colonic inflammation (Roager et al., 2018). This suggests that 3-IAA supplementation may be a therapeutic option for IBD patients.
Read the whitepaper: Learn more IBD through the lens of metabolomics in our whitepaper on the use of metabolomics to study complex chronic diseases.
Also in mice, supplementation with 3-IAA has been found to attenuate behaviors associated with depression and stress, enhance serotonin synthesis in the brain and gut, and alter gut microbiota (Chen et al., 2022).
Administration of 3-IAA via fecal transplant may be another route to influence gut microbiota in certain diseases. As discussed above, Tintelnot et al. demonstrated this in relation to cancer (Tintelnot et al., 2023). In another study, Ji et al. assessed the effects of 3-IAA in mice with high-fat diet-induced nonalcoholic fatty liver disease (NAFLD) (Ji et al., 2019). 3-IAA administered by intraperitoneal injection alleviated markers of insulin resistance and suggested a protective effect on liver damage.
While more research is needed, particularly in human subjects, these findings suggest promising avenues for exploring the use of 3-IAA as a biomarker and in therapeutic interventions.
References
Chen, Y. et al.: Indole Acetic Acid Exerts Anti-Depressive Effects on an Animal Model of Chronic Mild Stress. (2022) Nutrients | https://doi.org/10.3390/nu14235019
Chen, Y. et al.: Tryptophan-centered metabolic alterations coincides with lipid-mediated fungal response to cold stress. (2023) Heliyon | https://doi.org/10.1016/j.heliyon.2023.e13066
De Mello, M. et al.: Excited indole-3-aldehyde from the peroxidase-catalyzed aerobic oxidation of indole-3-acetic acid. Reaction with and energy transfer to transfer ribonucleic acid. (1980) Biochemistry | https://doi.org/10.1021/bi00564a019
Folkes, L. et al.: Oxidative activation of indole-3-acetic acids to cytotoxic species— a potential new role for plant auxins in cancer therapy. (2001) Biochemical Pharmacology | https://doi.org/10.1016/S0006-2952(00)00498-6
Gao, J.: Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. (2018) Front Cell Infect Microbiol. | https://doi.org/10.3389/fcimb.2018.00013
Gessner, A. et al.: A Metabolomic Analysis of Sensitivity and Specificity of 23 Previously Proposed Biomarkers for Renal Transporter-Mediated Drug-Drug Interactions. (2023) Clin Pharmacol Ther., | https://doi.org/10.1002/cpt.3017
Hendrikx et.al.: Indoles: metabolites produced by intestinal bacteria capable of controlling liver disease manifestation. (2019) J Intern Med. | https://doi.org/10.1111/joim.12892
Hishinuma, E. et al.: Wide-Targeted Metabolome Analysis Identifies Potential Biomarkers for Prognosis Prediction of Epithelial Ovarian Cancer. (2021) Toxins (Basel) | https://doi.org/10.3390/toxins13070461
Hou, Y. et al.: Tryptophan Metabolism and Gut Microbiota: A Novel Regulatory Axis Integrating the Microbiome, Immunity, and Cancer. (2023) Metabolites, | https://doi.org/10.3390/metabo13111166
Huang, Z. et al.: Impact of High-Fiber or High-Protein Diet on the Capacity of Human Gut Microbiota To Produce Tryptophan Catabolites. (2023) J Agric Food Chem., | https://doi.org/10.1021/acs.jafc.2c08953
Ji, Y. et al.: Indole-3-Acetic Acid Alleviates Nonalcoholic Fatty Liver Disease in Mice via Attenuation of Hepatic Lipogenesis, and Oxidative and Inflammatory Stress. (2019) Nutrients | https://doi.org/10.3390/nu11092062
Jones, L. et al.: Effects of indole-3-acetic acid on croton oil- and arachidonic acid-induced mouse ear edema. (1995) Inflammation Research | https://doi.org/10.1007/BF01797863
Krishnan, S. et al.: Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. (2018) Cell Rep. | https://doi.org/10.1016/j.celrep.2018.03.109
Lamas, B. et al.: CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. (2016) Nat | https://doi.org/10.1038/nm.4102
Lario, S. et al.: Plasma sample based analysis of gastric cancer progression using targeted metabolomics. (2017) Scientific Reports, | https://doi.org/10.1038/s41598-017-17921-x
Majima et al.: Synthetische Versuche in der Indol-Gruppe, VI.: Eine neue Synthese von β-Indolyl-alkylaminen. (1925) Berichte der Deutschen Chemischen Gesellschaft (A and B Series) | https://doi.org/10.1002/cber.19250580917
Mehta, O. et al.: Serum Metabolome Analysis Identified Amino-Acid Metabolism Associated With Pain in People With Symptomatic Knee Osteoarthritis – A Cross-Sectional Study. (2023) The Journal of Pain, | https://doi.org/10.1016/j.jpain.2023.02.023
Oluwagbemigun, K. et al.: Longitudinal relationship of amino acids and indole metabolites with long-term body mass index and cardiometabolic risk markers in young individuals. (2020) Scientific Reports | https://doi.org/10.1038/s41598-020-63313-z
Pavlova et al.: Urinary intermediates of tryptophan as indicators of the gut microbial metabolism. (2017) Anal Chimica Acta. | https://doi.org/10.1016/j.aca.2017.08.022
Riazati, N. et al.: Associations of microbial and indoleamine-2,3-dioxygenase-derived tryptophan metabolites with immune activation in healthy adults. (2022) Front. Immunol.| https://doi.org/10.3389/fimmu.2022.917966
Roager, H. et al.: Microbial tryptophan catabolites in health and disease.(2018) Nature Communications | https://doi.org/10.1038/s41467-018-05470-4
Rosas, H. et al.: A systems-level “misunderstanding”: the plasma metabolome in Huntington’s disease. (2015) Ann Clin Transl Neurol. | https://doi/10.1002/acn3.214
Russell, W. et al.: Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. (2013) Mol Nutr Food Res. | https://doi.org/10.1002/mnfr.201200594
Sadik, A. et al.: IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. (2020) Cell, | https://doi.org/10.1016/j.cell.2020.07.038
Shen, J. et al.: Indole-3-Acetic Acid Alters Intestinal Microbiota and Alleviates Ankylosing Spondylitis in Mice. (2022) Front. Immunol. | https://doi.org/10.3389/fimmu.2022.762580
Taleb, S.: Tryptophan Dietary Impacts Gut Barrier and Metabolic Diseases. (2019) Front Immunol. | https://doi.org/10.3389/fimmu.2019.02113
Tintelnot, J. et al.: Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. (2023) Nature | https://doi.org/10.1038/s41586-023-05728-y
Weissbach, H. et al.: Formation of Indole-3-acetic Acid and Tryptamine in Animals. (1959) Journal of Biological Chemistry | https://doi.org/10.1016/S0021-9258(18)70339-6
Wyatt et al.: Targeting Dietary and Microbial Tryptophan-Indole Metabolism as Therapeutic Approaches to Colon Cancer. (2021) Nutrients | https://doi.org/10.3390/nu13041189
Zhang, X. et al.: Endogenous Indole Pyruvate Pathway for Tryptophan Metabolism Mediated by IL4I1 (2020) J. Agric. Food Chem. | https://doi.org/10.1021/acs.jafc.0c03735