- History & Evolution
- Biosynthesis
- Bile acids and digestion
- Cholic acid in health and disease
- Cholic acid as a signaling molecule
- Cholic acid and the brain
- Cholic acid and the microbiome
- References
History & Evolution
1917: isolation and bulk production from ox bile for organic chemistry applications (Hofmann and Hagey 2014).
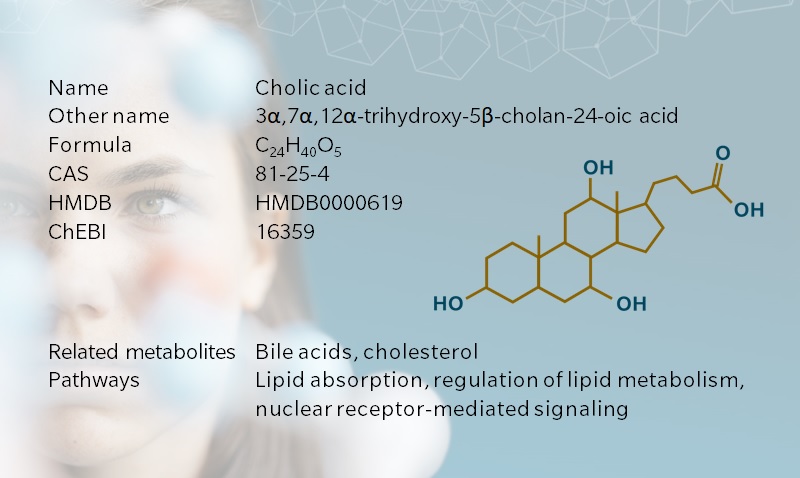
Cholic acid, from the Greek khole, meaning bile, is a primary bile acid synthesized in the liver of humans and other vertebrates. Together with other bile acids and bile salts, cholic acid contributes to the absorption of lipophilic nutrients in the intestine. Cholic acid also has a prominent role in the regulation of bile acid metabolism. As for many bile acids, the role of cholic acid in nuclear receptor-mediated signaling has become a hot topic in recent years, bringing new momentum to bile acid research.
Biosynthesis
In humans, cholic acid is synthesized from cholesterol in the liver by cytochromes P450 7A1, 8B1 and 27A1. Cholesterol is conjugated to glycine or taurine to form bile salts, which are secreted in the small intestine to aid the digestion and absorption of fat and other lipophilic nutrients. In the “classical pathway” of bile acid synthesis, cholic acid produces bile acid metabolites, leading to the production of deoxycholic acid (or DCA, a secondary bile acid), and glycine and taurine conjugates.
Bile acids and digestion
Conjugated primary bile acids (or bile salts) synthesized in the liver are stored in the gallbladder until needed in the intestine. Usually this is triggered by a large meal, which necessitates nutrient absorption in the colon. Once released in the intestine, primary conjugated bile acids are unconjugated from their glycine/taurine moieties and metabolized into secondary bile acids by the intestinal microbiota.
For cholic acid, this means that glycocholic acid (GCA) and taurocholic acid (TCA) are converted back to cholic acid before it is converted to the secondary bile acid deoxycholic acid (DCA). These secondary bile acids can in turn be conjugated to glycine/taurine (i.e. glycoldeoxycholic acid (GDCA) and taurodeoxycholic acid (TDCA) for DCA), to form amphipathic bile salts, which capture dietary fats and lipid-soluble nutrients to help their absorption through the enteric membrane.
Only a fraction of bile acids is estimated to be excreted in feces. The rest (both conjugated and unconjugated) are reabsorbed, returning to the liver via the enterohepatic circulation to be processed for reuse in digestion (Hofmann and Hagey 2014).
Cholic acid in health and disease
Cholic acid plays an important role in digestion, both as a precursor of several bile acids and bile salts, and as a regulator of bile acids and lipid metabolism. Mutations in the genes encoding enzymes involved in bile acid biosynthesis are rare, but result in serious pathologies involving the liver, kidneys and brain. They can also cause deficiencies due to the malabsorption of vitamins and other nutrients (Sundaram et al. 2008). Cholic acid has been successfully applied to the treatment of genetic diseases resulting in primary bile acid synthesis defects (Gonzales et al. 2018). As an inhibitor of bile acid synthesis (Lu et al. 2000), cholic acid has also been used to suppress this pathway in order to reduce the amount of toxic bile acids produced in the liver of patients with Zellweger spectrum disorders (Berendse et al. 2016).
Cholic acid as a signaling molecule
As molecular biology and genomics progressed, research has revealed that bile acids play an important role in gene and metabolism regulation. In 1999, researchers discovered their capacity to bind nuclear receptors and therefore regulate gene expression (Makishima et al. 1999; Parks et al. 1999; Wang et al. 1999). This allows bile acids to act on their own homeostasis (metabolism and transport), on cholesterol synthesis, and on energy metabolism from both lipids and glucose.
A notable bile acid receptor is the farnesoid X receptor (FXR), the first nuclear receptor found to mediate bile acid signaling. Interestingly, cholic acid was first thought to have no-to-low capacity for FXR activation. However, that assumption was based on experiments performed in cell models that did not express membrane transporters allowing the entry of hydrophilic bile acids, such as cholic acid. It was later demonstrated in other models that cholic acid was one of the bile acids that can activate FXR (Wang et al. 1999). In 2002, the G protein-coupled receptor, TGR5, expressed at the cell surface, was discovered as a mediator of bile acid signaling within cells (Maruyama et al. 2002). TGR5 activation can trigger anti-inflammatory signaling, influence liver and gallbladder functions, modulate thyroxine metabolism, and play a critical role in energy homeostasis (Duboc et al. 2014). Cholic acid is an agonist of TGR5, though less so than DCA or secondary bile acids (Kawamata et al. 2003).
Cholic acid and the brain
With their newly found effect as nuclear receptor ligands, bile acids have received a lot more attention in the neurology field in the last decade. Bile acids can act directly on receptors present in the brain, or can trigger signals via growth factors and hormones that act on the CNS (Mertens et al. 2017). Both conjugated and unconjugated bile acids have been shown to cross the blood-brain-barrier.
A recent study in rats concluded that cholic acid and other unconjugated bile acids are present in brain tissue at the same concentrations as in the circulation, suggesting passive diffusion of these lipophilic bile acids (Higashi et al. 2017). More hydrophilic forms are thought to be transported in the brain via active bile acid transporters similar to those found in other organs (Mertens et al. 2017). In 2019, metabolomics revealed that the bile acid profile in serum samples from patients with Alzheimer’s disease (AD) is associated with clinical biomarkers of AD (Nho et al. 2018). Cholic acid levels were found to be lower in AD patients, while the ratios of DCA to cholic acid, indicative of dysbiosis of the intestinal microbiome, was associated with cognitive decline (MahmoudianDehkordi et al. 2019).
Cholic acid and the microbiome
Knowing the importance of the microbiome in bile acid biosynthesis, it comes as no surprise that dysbiosis has a large impact on bile acid profiles, including the serum levels of primary bile acids (Semba et al. 2017). A recent study on the development of the gut microbiota in newborn mice showed that bile acid levels can drive the maturation of the microbiome (van Best et al. 2020). Although the bile acid profiles of humans and mice contain a few striking differences, the general patterns are similar, with cholic acid and chenodeoxycholic acid (CDCA) at the top of the same two main pathways (Li and Dawson 2019). Further studies on intestinal colonization in germ-free mice suggest a potential avenue for experiments on the effects of the microbiome on bile acid profiles (Wahlström et al. 2017).
Learn more about the roles of cholic acid and other bile acids in complex chronic diseases such as cancer, Alzheimer’s disease, depression, inflammatory bowel disease, multiple sclerosis and diabetes in our whitepaper “Complex chronic diseases have a common origin”.
References
Berendse K, Klouwer FCC, Koot BGP, Kemper EM et al.: Cholic acid therapy in Zellweger spectrum disorders (2016) Journal of inherited metabolic disease | https://doi.org/10.1007/s10545-016-9962-9
Duboc H, Taché Y, Hofmann AF: The bile acid TGR5 membrane receptor: From basic research to clinical application (2014) Digestive and Liver Disease | https://doi.org/10.1016/j.dld.2013.10.021
Gonzales E, Matarazzo L, Franchi-Abella S, Dabadie A et al.: Cholic acid for primary bile acid synthesis defects: a life-saving therapy allowing a favorable outcome in adulthood (2018) Orphanet journal of rare diseases | https://doi.org/10.1186/s13023-018-0920-5
Higashi T, Watanabe S, Tomaru K, Yamazaki W et al.: Unconjugated bile acids in rat brain: Analytical method based on LC/ESI-MS/MS with chemical derivatization and estimation of their origin by comparison to serum levels (2017) Steroids | https://doi.org/10.1016/j.bjpt.2017.07.001
Hofmann AF, Hagey LR: Key discoveries in bile acid chemistry and biology and their clinical applications: history of the last eight decades (2014) Journal of lipid research | https://doi.org/10.1194%2Fjlr.R049437
Kawamata Y, Fujii R, Hosoya M, Harada M et al.: A G Protein-coupled Receptor Responsive to Bile Acids* (2003) Journal of Biological Chemistry | https://doi.org/10.1074%2Fjbc.M209706200
Li J, Dawson PA: Animal models to study bile acid metabolism (2019) Biochimica et biophysica acta. Molecular basis of disease | https://doi.org/10.1016/j.bbadis.2018.05.011
Lu TT, Makishima M, Repa JJ, Schoonjans K et al.: Molecular Basis for Feedback Regulation of Bile Acid Synthesis by Nuclear Receptors (2000) Molecular cell | https://doi.org/10.1016/S1097-2765(00)00050-2
MahmoudianDehkordi S, Arnold M, Nho K, Ahmad S et al.: Altered bile acid profile associates with cognitive impairment in Alzheimer’s disease-An emerging role for gut microbiome (2019) Alzheimer’s & dementia : the journal of the Alzheimer’s Association | https://doi.org/10.1016/j.jalz.2018.07.217
Makishima M, Okamoto AY, Repa JJ, Tu H et al.: Identification of a nuclear receptor for bile acids (1999) Science (New York, N.Y.) | https://doi.org/10.1126/science.284.5418.1362
Maruyama T, Miyamoto Y, Nakamura T, Tamai Y et al.: Identification of membrane-type receptor for bile acids (M-BAR) (2002) Biochemical and Biophysical Research Communications | https://doi.org/10.1016/S0006-291X(02)02550-0
Mertens KL, Kalsbeek A, Soeters MR, Eggink HM: Bile Acid Signaling Pathways from the Enterohepatic Circulation to the Central Nervous System (2017) Frontiers in neuroscience | https://doi.org/10.3389/fnins.2017.00617
Nho K, Kueider-Paisley A, MahmoudianDehkordi S, Arnold M et al.: Altered Bile Acid Profile in Mild Cognitive Impairment and Alzheimer’s Disease: Relationship to Neuroimaging and CSF Biomarkers (2018) Alzheimer’s & dementia : the journal of the Alzheimer’s Association | https://doi.org/10.1016/j.jalz.2018.08.012
Parks DJ, Blanchard SG, Bledsoe RK, Chandra G et al.: Bile acids: natural ligands for an orphan nuclear receptor (1999) Science (New York, N.Y.) | https://doi.org/10.1126/science.284.5418.1365
Semba RD, Gonzalez-Freire M, Moaddel R, Trehan I et al.: Environmental Enteric Dysfunction is Associated with Altered Bile Acid Metabolism (2017) Journal of pediatric gastroenterology and nutrition | https://doi.org/10.1097/MPG.0000000000001313
Sundaram SS, Bove KE, Lovell MA, Sokol RJ: Mechanisms of disease: Inborn errors of bile acid synthesis (2008) Nature clinical practice. Gastroenterology & hepatology | https://doi.org/10.1038/ncpgasthep1179
van Best N, Rolle-Kampczyk U, Schaap FG, Basic M et al.: Bile acids drive the newborn’s gut microbiota maturation (2020) Nature communications | https://doi.org/10.1038/s41467-020-17183-8
Wahlström A, Kovatcheva-Datchary P, Ståhlman M, Khan M-T et al.: Induction of farnesoid X receptor signaling in germ-free mice colonized with a human microbiota (2017) Journal of lipid research | https://doi.org/10.1194/jlr.M072819
Wang H, Chen J, Hollister K, Sowers LC, Forman BM: Endogenous bile acids are ligands for the nuclear receptor FXR/BAR (1999) Molecular cell | https://doi.org/10.1016/S1097-2765(00)80348-2