- History & evolution
- Biosynthesis vs. dietary uptake
- Lactic acid and signaling
- Lactic acid and exercise
- Lactic acidosis and cancer
History & Evolution
1780: isolation from sour milk (Kompanje et al. 2007) | 1808: discovery that it is produced in muscles during exertion | 1895: start of industrial production of lactate
Lactic acid was first discovered in sour milk in 1780, by Swedish scientist Karl Wilhelm Scheele (Kompanje et al. 2007). Originally named “Mjölksyra,” which means “acid of milk” in Swedish, its current name is derived from the Latin word for milk, lactis. In the mid-19th century, German chemist Johann Joseph Scherer’s case reports described the presence of lactic acid in human blood under pathological conditions (Kompanje et al. 2007).
Araki and Zillessen discovered that lactic acid was produced when the oxygen supply to muscles in mammals and birds was interrupted, proving a link between tissue hypoxia and lactic acid production (Araki 1891) (Zillessen 1891).
In 1856, Louis Pasteur identified the involvement of Lactobacillus in the formation of lactic acid. The German pharmaceutical firm Boehringer Ingelheim used this method on an industrial scale from 1895, paving the way for large-scale manufacturing of lactic acid-based products for use in the food and textile industries (Ghaffar et al. 2014).
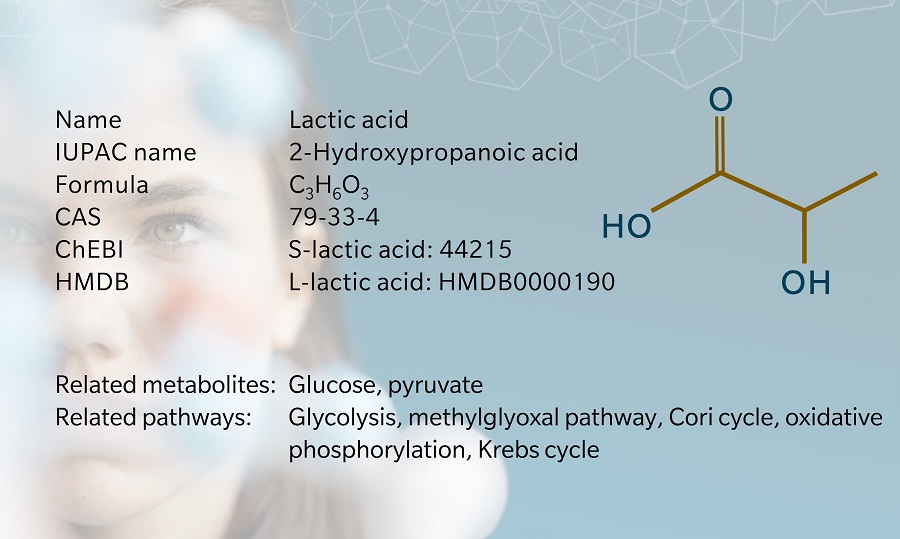
The terms “lactic acid” and “lactate” are frequently used interchangeably, but the metabolites are structurally different since lactic acid has an extra hydrogen atom compared to lactate. Lactic acid is unstable at physiological pH. As soon as it is produced, almost all of it is broken down into lactate and hydrogen at a pH of 7.35–7.45 (Sun et al. 2017)
Biosynthesis vs. dietary uptake
Lactate is the end product of anaerobic glycolysis, the final step of which is the conversion of pyruvate into lactate by the enzyme lactate dehydrogenase (LDH).
Because of an asymmetric carbon atom, lactate occurs in two stereoisomeric forms: L-lactate and D-lactate (Kowlgi 2015). Each is produced by a specific form of LDH (Farhana 2022). In humans, lactate is primarily produced as L-lactate. Lactate is transported through the plasma membrane thanks to several monocarboxylate transporters (MCTs) belonging to the SLC16 gene family (Halestrap 2013).
Lactic acid is produced by various cells in the body, including muscle cells, red blood cells and neurons. In an average adult, around 1,500 mM of lactic acid enters circulation every day (Sun et al, 2017). It’s mostly eliminated by the liver by incorporation into gluconeogenesis (via the Cori cycle) and ATP generation (via oxidative phosphorylation and the Krebs cycle).
Lactate is also found in various foods, particularly dairy products and fermented foods, such as pickled vegetables, cured meats and kimchi. Metabolomics studies have shed light on the fermentative capacity of lactic acid bacteria and the beneficial effects of the foods that contain lactic acid (Mozzi et al. 2013).
Lactic acid and signaling
Lactic acid is often considered the waste product of glycolysis. But as always with metabolism, there is no such thing as waste. In a fasted state, lactate can be taken up by other organs and continue its way to the tricarboxylic acid (TCA) cycle (Hui et al. 2017). In addition, lactate can serve as a substrate in signaling pathways.
Like many small molecules, lactic acid can also serve as a substrate to “decorate” other metabolites in a process called lactoylation (sometimes also called lactylation). Amino acids are the best described lactoylated metabolites. A similar process has been described for acetate, propionate, succinate, and other acyl groups linked to coenzyme A (CoA) and is perhaps best described in post-translational modifications (PTMs) of lysine residues on histones (Zhao et al. 2015).
Lactoylation of histones with a direct impact on gene expression is a recent discovery (Zhang et al. 2019). A commonly suggested donor group is lactoyl-CoA, as for other acyl-CoAs, but lactoyllysine has also been shown to be formed by the transfer of the lactoyl moiety from lactoylglutathione to lysine residues of proteins (Gaffney et al. 2020). This impacts protein function and has been described in histones and glycolytic enzymes, constituting a potent feedback mechanism to regulate glycolytic flux. In addition, a correlation of lysine acylations with the abundance of the corresponding acyl-CoA suggests another direct link to metabolism (Simithy et al. 2017).
Lactic acid and exercise
Muscles rely on oxygen to produce the energy they require during aerobic exercises, such as brisk walking, casual biking, or jogging. During strenuous, anaerobic exercise, like high-intensity interval training (HIIT), sprinting, weightlifting, and plyometrics, muscles generate this energy in the absence of oxygen. When muscles generate energy anaerobically, lactic acid is produced as a byproduct.
Athletes often associate lactic acid with a feeling known as “the burn”, which is common during anaerobic exercise. This was previously attributed to lactic acidosis – a build-up of excess lactic acid in the muscles. However, researchers have since discovered that the burning sensation is caused by elevated hydrogen ion levels triggering pain receptors in the muscles. This occurs when lactic acid breaks down into lactate, releasing hydrogen protons (Robergs 2004).
Interestingly, this metabolite may have played a role in the success of US Olympic swimmer Michael Phelps. He was found to have just 5.6 mM of lactate per liter of blood – about half to a third of that of his competitors – giving him an advantage in terms of endurance and recovery (Levine 2008).
Lactic acidosis and cancer
Lactic acidosis, the build-up of lactate in the body, results in an abnormally low blood pH level with several health consequences. It is usually caused by pre-existing acute or chronic medical conditions (such as heart failure, kidney disease or severe trauma (Bellomo 2002) (Adamo et al. 2017)), medications or poisoning. Symptoms may include nausea, vomiting, rapid and deep breathing, confusion, and extreme fatigue.
Cancer is frequently associated with lactic acidosis. In a process known as the Warburg effect, tumor cells produce energy through glycolysis, even in the presence of oxygen, resulting in lactic acid production. It is thought that cancer cells may suppress the immune response by maintaining a low pH level in their microenvironment (Choi et al. 2013).
Metabolomics may hold the key to characterizing the acidity of the tumor microenvironment. For example, metabolomics has shown that breast cancer cell lines respond differently to lactic acidosis based on their level of malignancy, a piece of information that can be used to tailor next-generation therapies (Gao et al. 2020).
Learn more about the roles of lactic acid and energy metabolism in complex chronic diseases such as cancer, Alzheimer’s disease, depression, inflammatory bowel disease, multiple sclerosis and diabetes in our whitepaper “Complex chronic diseases have a common origin”.
References
Adamo et al.: Prevalence of Lactic Acidaemia in Patients with Advanced Heart Failure and Depressed Cardiac Output (2017) Eur J Heart Fail | http://doi.org/10.1002/ejhf.628
Araki et al.: Ueber die Bildung von Milchsäure und Glycose im Organismus bei Sauerstoffmangel (1891) Z Physiol Chem | http://doi.org/10.1002/bchm1.1892.16.6.453
Bellomo et al.: Bench-to-Bedside Review: Lactate and the Kidney (2002) Crit Care | http://doi.org/10.1186/cc1518
Choi et al.: Cancer-generated Lactic Acid: a Regulatory, Immunosuppressive Metabolite? (2013) J Pathol | http://doi.org/10.1002/path.4218
Farhana et al.: Biochemistry, Lactate Dehydrogenase. In StatPearls [Internet] (2022) Treasure Island FL: StatPearls Publishing I https://www.ncbi.nlm.nih.gov/books/NBK557536/
Gaffney et al.: Non-enzymatic Lysine Lactoylation of Glycolytic Enzymes (2022) Cell Chem Biol | http://doi.org/10.1016/j.chembiol.2019.11.005
Gao et al.: Differential Metabolic Responses in Breast Cancer Cell Lines to Acidosis and Lactic Acidosis Revealed by Stable Isotope Assisted Metabolomics (2020) Nature | http://doi.org/10.1016/s41598-020-78955-2
Ghaffar et al.: Recent Trends in Lactic Acid Biotechnology: A Brief Review on Production to Purification (2014) Journal of Radiation Research and Applied Sciences | http://doi.org/10.1016/j.jrras.2014.03.002
Halestrap et al.: The SLC16 Gene Family – Structure, Role and Regulation in Health and Disease (2013) Mol Aspects Med | http://doi.org/10.1016/j.mam.2012.05.003
Hui et al.: Glucose Feeds the TCA Cycle via Circulating Lactate (2017) Nature | http://doi.org/10.1038/nature24057
Kompanje et al.: The First Demonstration of Lactic Acid in Human Blood in Shock by Johann Joseph Scherer (1814–1869) in January 1843 (2007) Intensive Care Med | http://doi.org/10.1007/s00134-007-0788-7
Kowlgi et al.: D-Lactic Acidosis: An Underrecognized Complication of Short Bowel Syndrome (2015) Gastroenterol Res Pract | http://doi.org/10.1155/2015/476215
Levine et al.: Out There (2008) New York Times | https://www.nytimes.com/2008/08/03/sports/playmagazine/803PHELPS-t.html
Mozzi et al.: Metabolomics as a Tool for the Comprehensive Understanding of Fermented and Functional Foods with Lactic Acid Bacteria (2013) Food Research International | http://doi.org/10.1016/j.foodres.2012.11.010
Robergs et al.: Biochemistry of Exercise-induced Metabolic Acidosis (2013) Am J Physiol Regul Integr Comp Physiol | http://doi.org/10.1152/ajpregu.00114.2004
Simithy et al.: Characterization of histone acylations links chromatin modifications with metabolism (2017) Nature Communications | http://doi.org/10.1038/s41467-017-01384-9
Sun et al.: Lactic Acid: No Longer an Inert and End-Product of Glycolysis (2017) American Journal of Physiology | http://doi.org/10.1152/physiol.00016.2017
Zhang et al.: Metabolic regulation of gene expression by histone lactylation (2019) Nature | https://doi.org/10.1038/s41586-019-1678-1
Zhao & Garcia: Comprehensive Catalog of Currently Documented Histone Modifications (2015) Perspectives in Biology | http://doi.org/10.1101/cshperspect.a025064
Zillessen et al.: Ueber die Bildung von Milchsäure und Glykose in den Organen bei gestörter Circulation und bei der Blausäurevergiftung (2017) Z Physiol Chem | http://vlp.mpiwg-berlin.mpg.de/references?id=lit16826