Non-response, loss of response and the need for stratification
Resistance to available drug therapies represents a major problem in pharmaceutical therapy. The issue affects a broad field of indications including (but not limited to) diabetes, depression and multiple sclerosis (Schork 2015). Non-response is not just an issue in clinical care, but is also a major obstacle in clinical trials for new pharmaceuticals, and may help explain why 50% to 80% of clinical development programs are discontinued at each stage of clinical trial (Pammolli et al. 2020).
In some disease groups, such as neurodegenerative diseases, failure at the clinical stage has unfortunately been the norm, with success stories a rare exception. Late-stage attrition may be driven by greater patient variability, due to different lifestyle factors, genetic backgrounds, comorbidities, and other factors. In this context, a higher proportion of patients must be expected to be therapy-resistant. Stratifying patients using novel biomarkers that reflect this complexity is likely to help differentiate the therapy-responsive patients and identify those who are likely to benefit from a particular therapeutic approach.
Stratification can be useful in other areas, too. In many diseases, therapies tend to become ineffective as the disease progresses, or because resistance mechanisms are activated. For example, acquired resistance is the subject of intense discussion in oncology (Floros et al. 2020, Bueschbell et al. 2022) and in relation to immunological therapies in inflammatory bowel disease (Fine et al., 2019). In Type 2 Diabetes, it is estimated that 5% to 10% of patients need to change therapy due to a loss of efficacy (Wexler et al. 2022).
Another significant challenge is the lack of actionable biomarkers that would enable clinical therapy management to optimize care with respect to co-medication and lifestyle therapies that could reduce non-response rates and improve clinical outcomes. Without such biomarkers, therapeutic choices must be made based on trial and error, although evidence in support of nutritional interventions is growing (Shastri et al. 2021, Schuetz 2017, Kaegi-Braun et al. 2021, Fan et al. 2022).
Figure 1: Areas of need for biomarker research
Why do metabolomic biomarkers hold promise?
Classical markers assessed in pharmaceutical research and development focus on concepts such as target engagement and monitoring of drug levels. In the clinical setting, only limited biomarkers are available, many of which are concentrated in the oncology space. Therapy choices are typically based on target (over-)expression of oncogenes (in “classical” targeted therapies) or of immune checkpoints (in cancer immunotherapies).
While such approaches are undoubtedly of great value, high rates of primary and/or acquired resistance remain a problem. This can be attributed to pathophysiological complexities that such markers do not properly consider. Tumor biology consists of much more than single oncogenic drivers, and the immune system is much too complex to determine by a single factor. (For more information about the pathophysiological process at play in oncology, Hanahan 2022 is a great choice.).
Metabolomics can help combat these issues in several ways. Comprehensive biochemical (i.e. metabolic) characterization of patients can reveal a lot of additional information about the various pathophysiological processes that contribute to the patient’s condition. These analyses can take account of the individual’s genetic background, age, comorbidities, and lifestyle factors, as well as interactions between organs that may contribute to the condition.
They can also be performed using easily accessible blood samples. Metabolomics can thus greatly enhance the information that is available from classical biomarker approaches and routine laboratory parameters. Finally, metabolic profiles are expected to evolve as the disease does, capturing the dynamics of the disease during a patient’s journey.
Consequently, metabolomics can provide actionable biomarkers that inform suitable approaches for co-medication and supportive nutrition therapy and can guide early adaptation of the therapeutic approach in the event of disease progression, development of acquired resistance, and/or toxicities.
A recent study has shown that biomarkers significantly improve the outcome of clinical trial success rates, particularly in the field of oncology (Parker et al. 2021). Exploratory biomarkers were found to bring a benefit to clinical trial success rates even before their proper validation.
The study did not refer to metabolomics specifically, but the factors discussed above suggest that the establishment of research programs for metabolomic biomarkers could constitute an opportunity for further improved success rates in clinical pharmaceutical research. As such, metabolic profiles may constitute a new cornerstone of precision therapy approaches.
Evidence for metabolic signatures as relevant and actionable biomarkers in pharmacological therapy
This discussion shows that in theory, metabolomics-based biomarker signatures have great potential. But is there proof that it works? According to several peer-reviewed papers, there is scientific evidence that it does.
Pharmacometabolomics in targeted cancer therapy
In a study of one of the most prominent examples of targeted cancer therapy, pharmacometabolomics showed promise as a tool for patient stratification in breast cancer patients treated with trastuzumab-paclitaxel (Miolo et al. 2015). While this treatment was an important milestone, but a relatively high proportion of recipients are non-responders.
The study showed that a simple ratio between spermidine and tryptophan is predictive of response. Spermidine interacts with pathways affected by the paclitaxel component of the treatment, while tryptophan is probably related to immunocytotoxicity of trastuzumab. Representing two chemically related and biophysically similar analytes (i.e. an amino acid and an amino acid metabolite), such ratios have the potential to serve as predictive biomarkers and could be easily implemented into clinical routine.
Several examples of translational biomarker research in targeted therapies have been published by the Cancer Therapeutics Unit at The Institute of Cancer Research in London by Dr. Raynaud and colleagues (Ang et al. 2016, Ang et al. 2017, Pal et al. 2020). As a similar experimental approach has been used in several projects, the results shall be discussed together. Figure 2 below shows that MEK inhibitors, PI3K inhibitors and AGC-kinase inhibitors act on interacting pathways. In mouse xenografts, differential dose-dependent metabolic responses have been observed.
The altered metabolites were considered candidate pharmacodynamic biomarkers, and the signals observed in preclinical research have largely been confirmed in Phase-I clinical trials. Moreover, the most important signals have associated with clinically relevant outcomes. In PI3K inhibitor therapy, branched-chain amino acids (BCAA) were associated with dose-limiting insulin resistance. In AGC-kinase inhibition, changes in nitric oxide (NO) metabolism were associated with dose-limiting hypotension. For MEK inhibitors, metabolic patterns were predictive of objective response and progression.
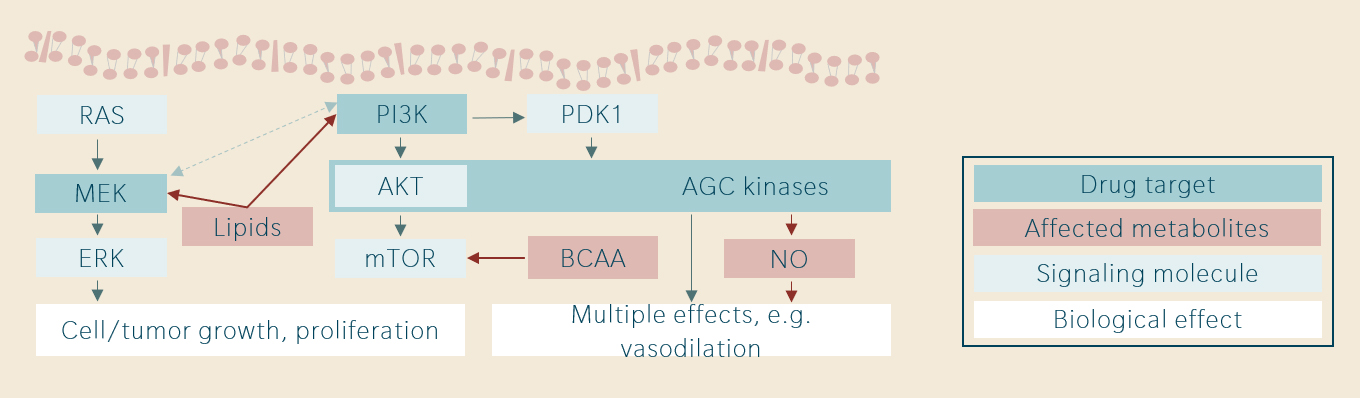
Figure 2: Signaling pathways, targets and metabolites (ICR London studies)
– Find out more about the AGC kinase inhibitor project and how circulating metabolites shed light on mechanism of action.
Pharmacometabolomics in cancer immunotherapy
Targeted therapies in oncology can have high initial response rates but high rates of acquired resistance. By contrast, immunotherapy often has long-lasting effects but only shows a response in a minority of patients. A group from the Heidelberg University Hospital in Germany found that lipids containing very long-chain fatty acids (VLCFA) were predictive to immunotherapy response (Mock et al. 2019). This suggests that supplementation with VLCFAs might increase a person’s response to immune checkpoint inhibition.
– For a deeper insight into this topic, take a look at our blog post: Make sure your drug fits to your cancer.
Tryptophan metabolism is a major regulator of immune processes. For this reason, indoleamine 2,3-dioxygenase (IDO) has attracted interest for its therapeutic potential. Although attempts to target IDO have largely failed thus far, the pathway remains of interest for the development of novel therapeutics (Peyraud et al. 2022).
Besides being a potential therapeutic target, the pathway also shows promise as a predictor of therapeutic outcomes. In a study of non-small cell lung cancer (NSCLC) patients, Kocher et al. (2021) describe an association between tryptophan metabolism and primary resistance to immune checkpoint inhibitors. Here, tryptophan is also suggested as a surrogate parameter for the IDO activity as a predictive biomarker for immune checkpoint inhibitor therapy, and as an informative trait for future investigations of therapeutic approaches targeting IDO directly.
Pharmacometabolomics in other indications
As noted, therapy resistance is a hot topic in virtually all areas of medicine, from neurodegenerative and neuropsychiatric diseases to cardiometabolic diseases. Beyond oncology, metabolomics also has proven potential as a stratification biomarker technology in other indications. For example, researchers from the Mayo Clinic in Rochester, US, have identified multiple genotype-metabolite interactions that are predictive of the response to antidepressant drugs escitalopram/citalopram (Joyce et al. 2021).
Selected acylcarnitines, lipids and amino acids showed pre-treatment differences between responders and non-responders. In addition, on-treatment changes in circulatory metabolite levels provided novel insights into the mechanism of action of those drugs, besides their actual target as selective serotonin reuptake inhibitors (SSRIs) (MahmoudianDehkordi et al. 2021).
Unsurprisingly, the prospect of improving therapeutic outcomes and patient stratification through metabolomics technologies has also attracted attention in the field of cardiometabolic diseases. For example, a research group around University Medical Center Groningen has found a signature consisting of 21 metabolites that predicts mircoalbuminuria as major endpoint of angiotensin II receptor blockers in patients with type 2 diabetes. The signature includes asymmetric dimethylarginine (ADMA), which may be related to the nitric oxide metabolism and endothelial function associated with the underlying pathophysiology (Pena et al. 2016).
Beyond stratification biomarkers, metabolomics offers significant benefits to pharmaceutical researchers through improved understanding of therapeutics’ mechanisms of action, and in the translation of results from discovery and preclinical research to clinical sciences.
– For more insights into the potential of pharmacometabolomics, see “Pharmacometabolomics provides biomarkers for precision medicine”.
References
Ang J. et al.: Plasma Metabolomic Changes following PI3K Inhibition as Pharmacodynamic Biomarkers: Preclinical Discovery to Phase I Trial Evaluation. (2016) Molecular Cancer Therapheutics | https://doi.org/10.1158/1535-7163.MCT-15-0815
Ang J. et al et al.: Modulation of Plasma Metabolite Biomarkers of the MAPK Pathway with MEK Inhibitor RO4987655: Pharmacodynamic and Predictive Potential in Metastatic Melanoma. (2017) Molecular Cancer Therapheutics | https://doi.org/10.1158/1535- 7163.MCT-16-0881
Bueschbell B. et al.: Network biology and artificial intelligence drive the understanding of the multidrug resistance phenotype in cancer. (2022) Drug Resistance Updatates | https://doi.org/10.1016/j.drup.2022.100811
Fan K. et al.: Targeting Nutrient Dependency in Cancer Treatment. (2022) Front. Oncol. | https://doi.org/10.3389/fonc.2022.820173
Fine S.: Etiology and Management of Lack or Loss of Response to Anti-Tumor Necrosis Factor Therapy in Patients With Inflammatory Bowel Disease. (2019) Gastroenterol Hepatol | https://pubmed.ncbi.nlm.nih.gov/31892912/
Floros K.: Investigating New Mechanisms of Acquired Resistance to Targeted Therapies: If You Hit Them Harder, Do They Get Up Differently? (2020) Cancer Research | https://doi.org/10.1158/0008-5472.CAN-19-3405
Hanahan D.: Hallmarks of Cancer: New Dimensions. (2022) Cancer Discov. | https://doi.org/10.1158/2159-8290.CD-21-1059
Joyce J. et al.: Multi-omics driven predictions of response to acute phase combination antidepressant therapy: a machine learning approach with cross-trial replication. (2021) Translational Psychiatry | https://doi.org/10.1038/s41398-021-01632-z
Kaegi-Braun N.: Association of Nutritional Support With Clinical Outcomes in Malnourished Cancer Patients: A Population-Based Matched Cohort Study. (2021) frontiers in Nutrition | https://doi.org/10.3389/fnut.2020.603370
Kocher F. et al.: High indoleamine-2,3-dioxygenase 1 (IDO) activity is linked to primary resistance to immunotherapy in non-small cell lung cancer (NSCLC). (2021) Translational Lung Cancer Research | https://doi.org/10.21037/tlcr-20-380
MahmoudianDehkordi S. et al.: Alterations in acylcarnitines, amines, and lipids inform about the mechanism of action of citalopram/escitalopram in major depression. (2021) Translational Psychiatry | https://doi.org/10.1038/s41398-020-01097-6
Miolo G. et al.: Pharmacometabolomics study identifies circulating spermidine and tryptophan as potential biomarkers associated with the complete pathological response to trastuzumab-paclitaxel neoadjuvant therapy in HER-2 positive breast cancer. (2016) Oncotarget | https://doi.org/10.18632/oncotarget.9489
Mock A.et al.: Serum very long-chain fatty acid-containing lipids predict response to immune checkpoint inhibitors in urological cancers. (2019) Cancer Immunological Immunotheraphy | https://doi.org/10.1007/s00262-019-02428-3
Pal A. et al.: Metabolomic changes of the multi (-AGC-) kinase inhibitor AT13148 in cells, mice and patients are associated with NOS regulation. (2020) Metabolomics | https//doi.org/10.1007/s11306-020-01676
Pammolli F. et al.: The endless frontier? The recent increase of R&D productivity in pharmaceuticals. (2020) Journal of Translational Medicine | https://doi.org/10.1186/s12967-020-02313-z
Parker J. et al.: Does biomarker use in oncology improve clinical trial failure risk? A large-scale analysis. (2021) Cancer Medicine | https://doi.org/10.1002/cam4.3732
Pena M.et al.: Serum metabolites predict response to angiotensin II receptor blockers in patients with diabetes mellitus. (2016) Journal of Transl ational Medicine | https://doi.org/10.1186/s12967-016-0960-3
Peyraud F. et al.: Targeting Tryptophan Catabolism in Cancer Immunotherapy Era: Challenges and Perspectives. (2022) froniers in Immunology | https://doi.org/10.3389/fimmu.2022.807271
Schork, N.: Personalized medicine: Time for one-person trials. (2015) Nature | https://doi.org/10.1038/520609a
Schuetz, P.: Food for thought: why does the medical community struggle with research about nutritional therapy in the acute care setting? (2017) BMC Med | https://doi.org/10.1186/s12916-017-0812-x
Shastri A. et al.: Personalized Nutrition as a Key Contributor to Improving Radiation Response in Breast Cancer. (2022) International Journal of Molecular Sciences | https://doi.org/10.3390/ijms23010175
Wexler, D. et al.: Management of persistent hyperglycemia in type 2 diabetes mellitus. (2022) Up to Date | https://www.uptodate.com/contents/management-of-persistent-hyperglycemia-in-type-2-diabetes-mellitus#H7554297


