History & evolution
Biosynthesis vs. dietary uptake
Histamine, the immune system, and allergies
Histamine, the gut, and the microbiome
Histamine and neurology
Histamine and cancer
History and evolution
1907: first synthesis from histidine (Tiligada et al. 2020) | 1910: first isolated from mold | 1937: synthesis of first antihistamine
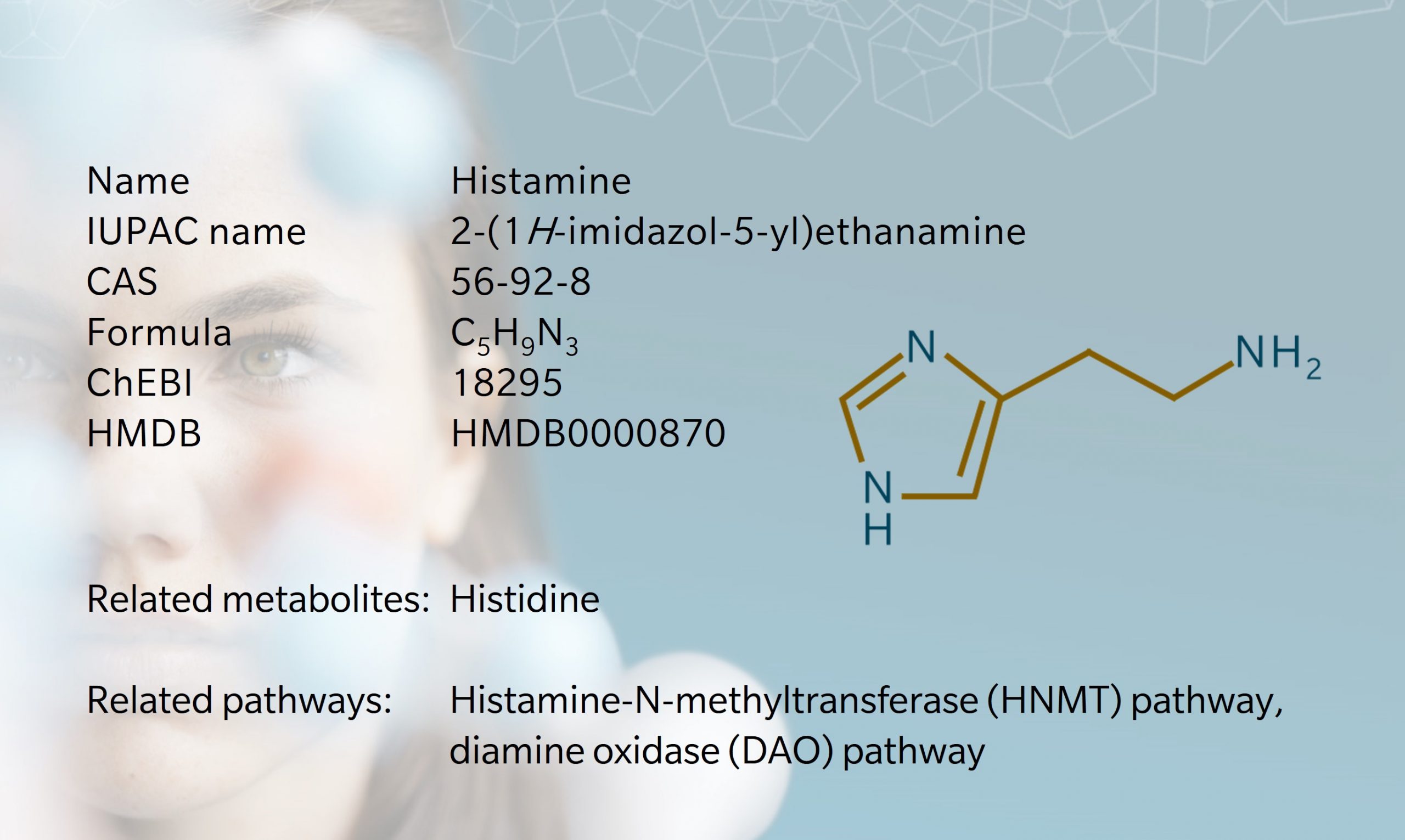
Histamine is an amine with multiple physiological effects, best known for its role in allergies and anaphylaxis. Histamine was first synthesized by Windhaus and Vogt in 1907, then isolated from mold by Dale and Laidlaw in 1910 (Dale et al. 1910). They referred to the substance as β-imidazolylethylamine, but it later became known as “histamine”, taking its name from the Greek word for tissue, histos (Tiligada et al. 2020). Histamine has since become one of the most well-researched substances in biomedical science, linked to multiple Nobel Prize awards. It’s also the only metabolite to have its very own anthem, courtesy of the European Histamine Research Society (EHRS, 2018).
Histamine is found in stinging nettles and insect venom, causing an itchy, painful reaction in anyone who is stung or bitten. In humans, early research focused on histamine’s role in the inflammatory response, particularly in local reactions to allergies. The discovery of antihistamines led to ground-breaking treatment for allergic disorders, and more recently in other immune-related disorders (Cataldi et al. 2014).
Histamine is found in nearly all tissues in the human body. It stimulates smooth muscle contraction, vasodilation, and gastric acid secretion, and plays a role in cell differentiation, proliferation and regeneration (Branco et al. 2018). In the brain, it acts as a neurotransmitter, carrying messages through the central nervous system (CNS). It has even been linked to sleep regulation, diabetes and obesity (Yoshimoto et al. 2006). Thus, histamine is a metabolite of interest in multiple disorders and potential treatments.
Biosynthesis vs. dietary uptake
Histamine is synthesized via the decarboxylation of the essential amino acid histidine. This occurs primarily in mast cells, but also in basophils throughout the body, histaminergic neurons in the brain, and enterochromaffin-like cells in the stomach (Huang et al. 2018). Basophils and mast cells can synthesize and store large amounts of histamine, whereas myeloid and lymphoid cells synthesize it without storing (Patel et al. 2022).
Histamine is stored in intracellular granules and released in response to different stimuli, such as immunoglobulin E (IgE) antibodies produced by the immune system. Degranulation is catalyzed by enzymes including diamine oxidase (DAO) and histamine-N-methyltransferase (HNMT). The DAO pathway converts histamine to imidazole-4-acetate and is associated with gastrointestinal activity. HNMT converts histamine to N-methylhistamine and acts in the CNS and airways. Around 50-80% of synthesized histamine is metabolized in the HNMT pathway, 15-30% in the DAO pathway, and 2-3% is excreted unchanged (Patel et al. 2022).
Once released, histamine binds to four different G protein-coupled receptors (GPCR), referred to as H1 to H4. Each one is activated in response to a different concentration of histamine, depending on where in the body the histamine is released (Panula et al. 2015). GPCRs form part of the largest family of membrane proteins in the human genome. Their structure includes an extracellular N terminus, intracellular C terminus and seven transmembrane helices joined by three intracellular and three extracellular loops (Panula et al. 2015).
The expression of these receptors and their response to histamine is as follows :
- H1-Receptors (H1R) are expressed in multiple cells and tissues in response to endogenous histamine. They play a key role in allergy and inflammation. Stimulation causes vasodilation, bronchoconstriction, mucus secretion and platelet activation (Thangam et al. 2018).
- H2-Receptors (H2R) are also widely expressed, and when activated can cause excess mucus in the airways, vascular permeability, increased heart rate and gastric acid secretion (Thangam et al. 2018).
- H3-Receptors (H3R) are found in the nervous system, regulating histamine by inhibiting histamine synthesis. H3R are involved in blood-brain barrier function, sleep cycles, cognitive function, homeostatic energy regulation and neurotransmission (Yoshimoto et al. 2006).
- H4-Receptors (H4R) are expressed in mast cells, particularly in bone marrow, white blood cells and the oral epithelium. While H3R inhibits histamine production, H4R stimulates histamine and cytokine production. H4R was discovered via the human genomic DNA database in 2000 (Oda et al. 2000).
Histamine can also be synthesized from bacteria in food, such as histidine decarboxylases found in lactic acid bacteria in fermented food (Landete et al. 2008). Histamine-rich foods include alcohol, dairy products, fermented foods and certain vegetables (Reese et al. 2018).
Histamine, the immune system, and allergies
Histamine’s role in allergic reactions was first discovered in 1932, and we now know it plays a central role in autoimmune conditions, inflammation and inflammatory disease (Patel et al. 2022). It has both pro- and anti-inflammatory effects, depending on which receptor is activated and in which cells. In the event of local tissue damage or infection, the release of histamine causes vasodilation, which allows leukocytes and immunoproteins to move easily to the damaged site and fight infection.
While this immune response is often helpful, hypersensitivity (as in hay fever, asthma or allergies) can cause unnecessary and unpleasant symptoms, such as sneezing, wheezing, itching, swelling, increased heart rate, eye irritations and gastric issues (Thangam et al. 2018). In extreme cases, anaphylaxis occurs, which can be fatal.
Antihistamines work by preventing histamine from binding to the relevant receptor, thus inhibiting the inflammatory response (Thangam et al. 2018). Common antihistamines to treat allergies are H1 antagonists. Antihistamines that block H2R inhibit gastric acid secretion, and would include ulcer treatments.
Histamine, the gut, and the microbiome
High concentrations of histamine are found in the gastrointestinal (GI) tract, so it’s no surprise that this metabolite is involved in many GI processes and disorders. However, there remain gaps in our understanding of how microbiota mediate histamine and histamine receptor activity in the gut. Enzymatic activity, dietary intake and microbial processes can all affect histamine levels. While it can have a protective effect, excess levels of histamine are linked to several mucosal inflammatory disorders, including food allergies, histamine intolerance, irritable bowel syndrome and inflammatory bowel disease (Smolinksa et al. 2013).
More than 20% of the population suffer from gastrointestinal problems (Schnedl et al. 2021). Unfortunately, the etiology and pathophysiology of many of these disorders remains unclear. The DAO pathway is the subject of much interest, given its role in breaking down ingested histamine.
A deficiency of DAO leads to impaired histamine degradation, which causes histamine intolerance (HIT). People with HIT suffer a variety of adverse reactions when they consume histamine-rich foods. This should not be confused with food allergies, though histamine often plays a role there too. Enterochromaffin cells have been shown to affect intestinal food intolerances associated with excessive histamine (Pfangazl et al. 2019).
DAO activity has been found to correspond to mucosal damage in Crohn’s disease, ulcerative colitis and colonic adenoma (Schnedl et al. 2021). A study of patients with gastric cancer found that DAO may be a useful predictor of GI toxicity in response to anticancer drugs (Miyoshi et al. 2015). Histamine also attracts interest as a potential therapeutic target in irritable bowel syndrome (Fabisiak et al. 2017).
Histamine and neurology
In the brain, histamine’s effects are mediated by all four receptors (Panula et al. 2021). H1R and H2R are involved in the wake-sleep cycle. H3R regulates neurotransmission, including serotonin- and glutamate-mediated. H4R is present in cerebral blood vessels and microglia, though its exact role remains undefined. Microglial modulation may be a therapeutic target for disorders such as Parkinson’s disease and multiple sclerosis, where changes have been found in histamine receptor levels (Iida et al. 2022).
High levels of histamine are found in the basal ganglia of people with Parkinson’s disease, with increased concentrations in blood and cerebrospinal fluid (Ayaz et al. 2022). Because histamine receptors play a role in motor functioning, they are a potential therapeutic target (Fang et al. 2021). Studies have shown that antihistamines can have a positive effect on resting tremors. However, first generation antihistamines have also been associated with an increased risk of dementia, which suggests they are not a suitable treatment (Gray et al. 2015).
H4R ligands may have a role in the treatment of neuroimmunological disorders and inflammatory neurodegenerative disorders, though more research is needed (Panula et al. 2021).
Histamine dysregulation is also associated with neuropsychological disorders including Tourette’s syndrome, autism spectrum disorders, attention deficit hyperactivity disorder and schizophrenia (Carthy et al. 2021).
Histamine and cancer
Histamine’s effect on the regulation and behavior of different immune cells draws attention in the world of oncology. Studies have shown that histamine directly influences carcinogenesis, and thus may be a potential therapeutic target (Sun et al. 2018).
A spatially resolved metabolomics method was used to analyze tumor-associated metabolite and enzyme changes in esophageal cancer tissues (Sun et al. 2018). Histamine was significantly downregulated in cancer tissue, and decarboxylation of histidine was found to be weaker in cancer tissue than in non-cancerous tissues.
Antihistamines have also been associated with improved clinical outcomes in cancer treatment. High histamine levels seem to inhibit the immunotherapy response, suggesting a role for antihistamines as an adjuvant treatment (Hongzhong et al. 2022).
References
Ayaz et al.: Parkinsonism Attenuation by Antihistamines via Downregulating the Oxidative Stress, Histamine, and Inflammation. (2022) ACS Omega | https://doi.org/10.1021/acsomega.2c00145.
Branco et al.: Role of Histamine in Modulating the Immune Response and Inflammation. (2018) Mediators Inflamm | https://doi.org/10.1155/2018/9524075.
Carthy et al.: Histamine, Neuroinflammation and Neurodevelopment: A Review. Front. (2021) Neurosci | https://doi.org/10.3389/fnins.2021.680214.
Cataldi et al.: Histamine receptors and antihistamines: from discovery to clinical applications. (2014) Chem Immunol Allergy | https://doi.org/10.1159/000358740.
Dale et al.: The physiological action of β-iminazolylethylamine. (1910) J Physiol | https://doi.org/10.1113/jphysiol.1910.sp001406.
EHRS (2018) European Histamine Research Society | https://www.ehrs.org.uk.
Fabisiak et al.: Targeting Histamine Receptors in Irritable Bowel Syndrome: A Critical Appraisal. (1917) J Journal of Neurogastroenterology and Motility | https://doi.org/10.5056/jnm16203.
Fang et al.: Histamine-4 receptor antagonist ameliorates Parkinson-like pathology in the striatum. (2021) Brain, Behavior, and Immunity | https://doi.org/10.1016/j.bbi.2020.11.036 .
Gray et al.: Cumulative use of strong anticholinergics and incident dementia: a prospective cohort study. (2015) JAMA Intern Med | https://doi.org/10.1001/jamainternmed.2014.7663.
Hongzhong et al.: The allergy mediator histamine confers resistance to immunotherapy in cancer patients via activation of the macrophage histamine receptor H1. (2022) Cancer Cell | https://doi.org/10.1016/j.ccell.2021.11.002
Huang et al.: Molecular Regulation of Histamine Synthesis. (2018) Front Immunol | https://doi.org/10.3389/fimmu.2018.01392
Iida et al.: Histamine and Microglia. (2022) Curr Top Behav Neurosci | https://doi.org/10.1007/7854_2022_322
Landete et al.: Updated molecular knowledge about histamine biosynthesis by bacteria. (2008) Crit Rev Food Sci Nutr | https://doi.org/10.1080/10408390701639041
Miyoshi et al.: Serum diamine oxidase activity as a predictor of gastrointestinal toxicity and malnutrition due to anticancer drugs. (2015) Journal of Gastroenterology and Hepatology | https://doi.org/10.1111/jgh.13004
Oda et al.: Molecular Cloning and Characterization of a Novel Type of Histamine Receptor Preferentially Expressed in Leukocytes. (2000) Journal of Biological Chemistry | https://doi.org/10.1074/jbc.M006480200.
Panula et al.: International Union of Basic and Clinical Pharmacology. XCVIII. Histamine Receptors. (2015) Pharmacol Rev | https://doi.org/10.1124/pr.114.010249.
Panula et al.: Histamine receptors, agonists, and antagonists in health and disease. (2021) Handb Clin Neurol | https://doi.org/10.1016/B978-0-12-820107-7.00023-9.
Patel et al.: Biochemistry, Histamine. (2022) In: StatPearls [Internet]. Florida: StatPearls Publishing | https://www.ncbi.nlm.nih.gov/books/NBK557790
Pfangazl et al.: Histamine causes influx via T-type voltage-gated calcium channels in an enterochromaffin tumor cell line: potential therapeutic target in adverse food reactions. (2019) Am J Physiol Gastrointest Liver Physiol | https://doi.org/10.1152/ajpgi.00261.2018
Reese et al.: Nutrition therapy for adverse reactions to histamine in food and beverages. (2018) Allergol Select | https://doi.org/10.5414/ALP34152
Schnedl et al.: Histamine Intolerance Originates in the Gut. (2021) Nutrients | https://doi.org/10.3390/nu13041262
Sun et al.: Spatially resolved metabolomics to discover tumor-associated metabolic alterations. (2018) PNAS | https://doi.org/10.1073/pnas.1808950116
Smolinska et al.: Histamine and gut mucosal immune regulation. (2013) Allergy | https://doi.org/10.1111/all.12330
Thangam et al.: The Role of Histamine and Histamine Receptors in Mast Cell-Mediated Allergy and Inflammation: The Hunt for New Therapeutic Targets. (2018) Front Immunol | https://doi.org/10.3389/fimmu.2018.01873
Tiligada et al.: Histamine pharmacology: from Sir Henry Dale to the 21st century. (2020) Br J Pharmacol | https://doi.org/10.1111/bph.14524
Yoshimoto et al.: Therapeutic potential of histamine H3 receptor agonist for the treatment of obesity and diabetes mellitus. (2006) PNAS | https://doi.org/10.1073/pnas.0506104103